GrindBio is a protein company, and we think a lot about how to properly express proteins in E. coli. We like E. coli because its internals are pretty well known. Researchers have been toying with E. coli, pulling its strings and kicking its flagella for decades. Also, Cheap. Getting E. coli to express a protein is like sitting underneath a money tree and having the tiny waterlords rake it for you. So, we like to start with E. coli because everyone starts here.
E. coli is a living organism, and even though its genome is 4.5Mb, about a thousandth of the human genome, it’s still complex.1 Sometimes, inserting a human gene into E. coli and getting it to express properly is straightforward. However, more often than not, various variables need to be optimized, which take time and effort.
What are a few of the variables?
There’s fusion tags. There’s codon optimization. There’s E. coli strain. There’s temperature and co-expressing chaperones. There’s promoters and inducers and concentrations. Any, or all of these, can be tested. The complexity can easily overwhelm 2.

There are so many variables in E. coli expression that being aware of them can be challenging. And being aware of the latest research for each one of the variables is also challenging. What keeps me awake at night, is the idea that the solution to an insoluble3 protein is just that I haven’t found the right conditions for each of the variables4 and I need to do more experimenting.
And experimenting with each of the variables is hard to do.5
The impact and play of multiple variables working together to enhance the soluble expression of a heterologous gene in E. coli is called the expression space6. A 3D surface plot shows what an expression space might look like: peaks, valleys, smooth regions, bumpy areas.
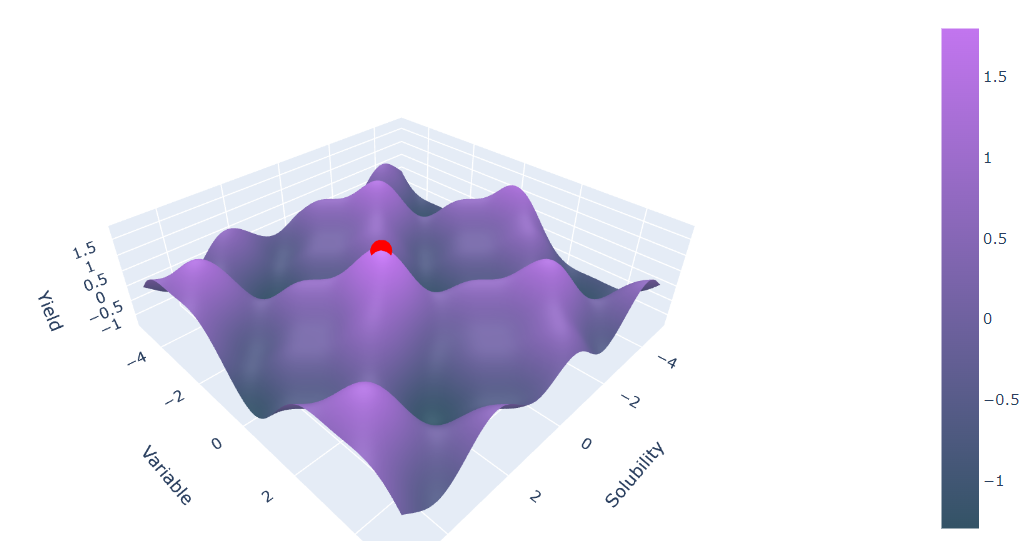
Notice the axes? For E. coli, the yield is a function of solubility and variable. By systematically exploring the expression space, we can identify the best conditions for high protein yield (the peaks), leading to more efficient and cost-effective production.7
Finding those peaks is what E. coli expression is all about. And there’s no better feeling than finding a peak.
And that’s the novelty of GrindBio: we test multiple variables simultaneously hunting for expression peaks.
Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The Pangenome Structure of Escherichia coli: Comparative Genomic Analysis of E. coli Commensal and Pathogenic Isolates. J Bacteriol 190: https://doi.org/10.1128/jb.00619-08 ↩
Consider 4 conditions for each variable and 8 variables. The number of experiments is (8^4) 4096. This doesn't even consider replicates. Put that in your incubation chamber and shake it! ↩
A protein that forms inclusion bodies or produces a low soluble yield. ↩
As a graduate student, I probably should have stopped experimenting with all the variables in E. coli and moved on to another system, but I was tantalized by the idea that I could easily change a single variable like the E. coli strain and it would express for me. So, I continued testing well beyond sanity. ↩
There are two outcomes to testing variables: 1) The protein becomes more soluble and the variable can be further explored for greater expression. 2) The variable does not affect expression, and another variable needs to be selected for testing. It’s more common to discover that a single variable does not affect expression. ↩
I love that word. It works with so many other subject areas. Like, what’s the expression space for the perfect batch of cookies? Or, many musicians playing or singing a piece with slight variations can be the expression space of a song. The expression space of successful marriages is a function of the individual traits of the couple and their environment. It’s broader for some couples, smaller for others. ↩
Imagine an experiment with a concentration of 0.7 mM inducer performed at a temperature of 37°C. Keep the inducer concentration the same but decrease the temperature to 18°C. That’s two points in our expression space. Maybe we get more protein at 37°C because E. coli likes that temperature and inducer concentration, and there’s a high protein yield as a result. At 18°C the expression isn’t as plentiful. Imagine trying several other inducer concentrations: 0.01 all the way to 1.5 mM separated by small intervals because we’re curious. We find that the best protein yield occurs at 0.7 mM. On our expression space topology, a peak can be found for solubility and inducer concentration at 0.7. ↩